Title: Characterisation of the kisspeptin system: The role of sex, obesity, and endocrine disruptors
Title: Brain-Derived Neurotrophic Factor (BDNF): Interactions with the serotonergic system and its potential as a biomarker in neurological and neuropsychiatric diseases
Title: Serotonin receptor studies in the pig brain: pharmacological intervention and positron emission tomography tracer development
Title: Molecular neuroimaging of synaptic plasticity in the human brain
Title: A Pig Model for Studies of Serotonergic Mechanisms in Memory Disorders
Title: Changes in the 5-HT4 receptor in animal models of depression and antidepressant treatment
Title: Towards personalized medicine Effectiveness of pretreatment EEG biomarker in Major Depressive Disorder
Title: Critical Investigations of Two Meditation Based Stress Reduction Programs and of Mindfulness as a Predictor of Mental Health in the Population
Title: Proprioception - an obstacle for motor control in conditions with a visuoproprioceptive conflict
Title: In Vivo Serotonergic Markers in Overweight and Schizophrenic Human Subjects
Title: Pharmacokinetic and pharmacodynamic evaluation of classical psychedelic substances in the human brain
Title: Development and Evaluation of Potential 5-HT7 Receptor PET Tracer Candidates
Title: Evaluation of PET radioligands for the cerebral 5-HT7 and 5-HT2A receptors
Title: Frontal Dopamine D2/3 Receptors and Brain Structure in Antipsychotic-Naïve Schizophrenia Patients Before and After Their First Antipsychotic Treatment
Title: Exploring nicotinic receptors and histone deacetylases through neuroimaging
Title: PET Imaging of Cerebral Serotonin 4 Receptors in Relation to Sex Aging and Alzheimer's Disease
Title: Neurobiological Substrates of Depression and Treatment Response - From Brain Structure to Sex Hormones
Title: The serotonin 4 receptor binding as a novel imaging marker in major depressive disorder and the association to antidepressant treatment response
Title: Molecular Brain Imaging of the Serotonin System: Reproducibility and Evaluation of PET Radiotracers
Title: I should have been a bear. Bears are allowed to hibernate; humans are not
Title: Characterization of the alpha7 nicotinic receptor and Lynx proteins and their relation to Alzheimer's desease - a translational neurobiology study
Title: PET investigations of brain serotonin receptor binding in migraine patients
Title: The role of serotonin 2A receptors in prefrontal cortex function - Implications in schizophrenia
Title: In Vivo PET Imaging of the Cerebral 5-HT4 Receptors in Healthy Volunteers - In Relation to Appetite, Memory and Pharmacological Intervention
Title: Epilepsy surgery: Outcomes of the Danish evaluation program and development of new EEG based methods
Title: Evaluation of the superselective radioligand [123I]PE2I for imaging of the dopamine transporter
in SPECT
Title: Translocator Protein Imaging with 123I-CLINDE SPECT - Method Development and Clinical Research
Title: Corrections in clinical Magnetic Resonance Spectroscopy and SPECT - Motion correction in MR spectroscopy Downscatter correction in SPECT
Title: Homeostatic sleep mechanisms and brain-fluid dynamics in humans
Title: Genetic and Epigenetic Determinants of Serotonin Neurotransmission: Mapping Predictive Risk- and Treatment Markers for Depressive Episodes
Title: Imaging brain serotonin 2A receptors: Methodological and genetic aspects and involvement in Tourette’s syndrome
Title: Biological aspects of postpartum mental health: prevention opportunities and mother’s perceptions
Title: Hormonal contributions to depressive episodes in women - Insights from register-based cohort studies from Denmark
Title: Relating cerebral serotonin 2A receptor and serotonin transporter binding to personality and familial risk for mood disorder
Title: Shining a light on the black cloud of depression - A study of cognitive markers in Major Depressive Disorder
Title: Brain-Derived Neurotrophic Factor (BDNF) and glucocorticoids: Influence on serotonin 2A receptors and relation to major depression
Title: Functional and Molecular Imaging of the Serotonin System in the Human Brain
Description
Figure 1. Average in vivo SV2A density maps (Bmax; pmol/mL) on the FreeSurfer fsaverage surface (top) and in the MNI152 volume space (bottom).
Publication
Johansen A, Beliveau V, Colliander E, Raval NR, Dam VH, Gillings N, Aznar S, Svarer C, Plavén-Sigray P, Knudsen GM. An In Vivo High-Resolution Human Brain Atlas of Synaptic Density. J Neurosci. 2024 Aug 14;44(33):e1750232024
Downloads
Average, standard deviation (SD) and coefficient of variation (COV) density (Bmax) maps on the FreeSurfer surface (fsaverage) and in the MNI152 space.
Note: Cortical data is mapped in the MNI152 space for visualization purposes only.
Contacts
For questions related to the NRU SV2A atlas, contact Annette Johansen or Vincent Beliveau.
Copyright
Copyright (c) 2023, Neurobiology Research Unit, Copenhagen University Hospital, Rigshospitalet.
All rights reserved according to CC 4.0 BY-NC-SA (https://creativecommons.org/licenses/by-nc-sa/4.0).
Official guidelines when applying for data from the Cimbi database
Application form to gain official access to data from the Cimbi database
Description
Serotonin (5-HT) is a key neurotransmitter involved in a broad range of brain functions and behaviors and is implicated in the pathophysiology of neuropsychiatric illnesses. Here, we present for the first time a PET and MR-based high resolution atlas of some main 5-HT receptors (5-HT1AR, 5-HT1BR, 5-HT2AR and 5-HT4R) and the 5-HT transporter, generated from 210 healthy individuals scanned with different selective PET-radioligands. We related our molecular imaging atlas to seminal autoradiography data and found an unprecedented agreement, supporting the validity of the methodology and results presented here, and allowing us to translate PET binding estimates into densities. As such, this conversion facilitates the interpretability of the atlas and allows for a direct comparison across the five 5-HT targets, in vivo in the human brain. Furthermore, we compare for the first time in vivo in humans the regional target densities to mRNA levels. Our results highlight intrinsic properties of the various 5-HT receptors, furthering the understanding of their individual contribution to the 5-HT system. Taken together, these findings provide novel insights into fundamental properties of the 5-HT system.
Data
All participants included in this study were healthy controls from the Cimbi database (Knudsen et al., 2015); the data analysis was restricted to include individuals aged between 18 and 45 years. Participants were recruited by advertisement for different research protocols approved by the Ethics Committee of Copenhagen and Frederiksberg, Denmark. A total of 232 PET scans and corresponding structural MRI were acquired for 210 individual participants; 189 subjects had only 1 scan, 20 subjects had 2 scans and a single had 3 scans.
Methods
PET data was acquired in list-mode on a Siemens HRRT scanner operating in 3D-acquisition mode, with an approximate in-plane resolution of 2 mm (1.4 mm in the center of the field of view and 2.4 mm in cortex) (Olesen et al., 2009). PET frames were reconstructed using a 3D-OSEM-PSF algorithm (Comtat et al., 2008; Sureau et al., 2008). Scan time and frame length were designed according to the radiotracer characteristics. Dynamic PET frames were realigned using AIR 5.2.5 (Woods et al., 1992).
T1 and T2-weighted structural MRI were acquired on four different Siemens scanners with standard parameters. All structural MRIs (T1 and T2) were unwarped offline using FreeSurfer's gradient_nonlin_unwarp v0.8 or online on the scanner (Jovicich et al., 2006). For further details on structural MRI acquisition parameters, see Knudsen et al. (2016).
Atlas maps
Brain regional BPND values were compared to the corresponding receptor density measurements from post-mortem autoradiography data from Varnäs et al. (2004) and Bonaventure et al. (2000) (for 5-HT4R). For all five targets, we found good to excellent associations between BPND and Bmax. The slope estimates of the regression were used to transform the BPND atlases into Bmax atlases (Figures 1 and 2), allowing for a direct comparison across targets. The regional densities are presented in Figure 3. No global or regional significant effect of age, gender or age x gender was found.
Figure 1. Average density (Bmax) maps for five 5-HT targets on the common FreeSurfer surface (left hemisphere; lateral view, upper and medial view, lower). Color scaling was individually adjusted in order to highlight features of the distributions.
Figure 2. Average density (Bmax) maps for the five 5-HT targets in the common MNI152 space (coronal, upper, z=8mm and sagittal, lower, x=-3mm). Color scaling was individually adjusted to highlight features of the distributions.
Figure 3. Density values (Bmax) of the five 5-HT targets in FreeSurfer defined brain regions. Median raphe is not reported for 5-HTT due the irreversible kinetic of the TACs, see also the Material and Methods section. A table with Bmax values for all regions can be downloaded below.
Clustering of atlas maps
The abovementioned human brain atlas of the serotonin (5-HT) system does not conform with commonly used parcellations of neocortex, since the spatial distribution of homogeneous 5-HT receptors and transporter is not aligned with such brain regions. This discrepancy indicates that a neocortical parcellation specific to the 5-HT system is needed. Hence we present parcellations of the 5-HT system created using a clustering approach focused on identifying stable and homogeneous clusters and derived from brain MR- and high-resolution PET images of five different 5-HT targets from 210 healthy controls. This is the same data that was used in the derivation of the atlas above. We then explore how well this new 5-HT parcellation can explain mRNA levels of all 5-HT genes. The parcellation derived here represents a characterization of the 5-HT system which is more stable and explains the underlying 5-HT molecular imaging data better than other atlases, and may hence be more sensitive to capture region-specific changes modulated by 5-HT.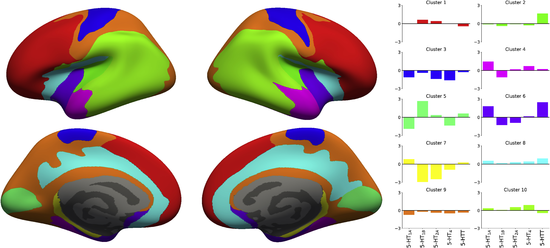
Figure 4. Parcellation obtained with K = 10 clusters and the associated regional 5-HT profile for each region. The parcellation is presented on the inflated fsaverage surface medial (lower) and lateral (upper) for both hemisphere (left and right).
Publications
The following publications should be referenced when using this atlas:
A high-resolution in vivo atlas of the human brain's serotonin system
Vincent Beliveau, Melanie Ganz, Ling Feng, Brice Ozenne, Liselotte Højgaard, Patrick M. Fisher, Claus Svarer, Douglas N. Greve, Gitte M. Knudsen
J Neurosci. 2017 Jan 4; 37(1): 120 - 128.
The Structure of the Serotonin System: a PET Imaging Study
Vincent Beliveau, Brice Ozenne,Stephen Strother, Douglas N. Greve, Claus Svarer, Gitte M. Knudsen, Melanie Ganz
Neuroimage. 2020 Jan 15; 205:116240
Bibliography
Bonaventure P, Hall H, Gommeren W, Cras P, Langlois X, Jurzak M, Leysen JE (2000) Mapping of serotonin 5-HT(4) receptor mRNA and ligand binding sites in the post-mortem human brain. Synapse 36:35-46.
Comtat C, Sureau FC, Sibomana M, Hong IK, Sjoholm N, Trebossen R (2008) Image based resolution modeling for the HRRT OSEM reconstructions software. In: 2008 IEEE Nuclear Science Symposium Conference Record, pp 4120-4123. IEEE.
Jovicich J, Czanner S, Greve D, Haley E, van der Kouwe A, Gollub R, Kennedy D, Schmitt F, Brown G, Macfall J, Fischl B, Dale A (2006) Reliability in multi-site structural MRI studies: effects of gradient non-linearity correction on phantom and human data. Neuroimage 30:436-443.
Knudsen GM et al. (2016) The Center for Integrated Molecular Brain Imaging (Cimbi) Database. Neuroimage:1-7.
Olesen OV, Sibomana M, Keller SH, Andersen F, Jensen J, Holm S, Svarer C, Højgaard L (2009) Spatial resolution of the HRRT PET scanner using 3D-OSEM PSF reconstruction. IEEE Nucl Sci Symp Conf Rec:3789-3790.
Sureau FC, Reader AJ, Comtat C, Leroy C, Ribeiro M-J, Buvat I, Trébossen R (2008) Impact of image-space resolution modeling for studies with the high-resolution research tomograph. J Nucl Med 49:1000-1008.
Varnäs K, Halldin C, Hall H (2004) Autoradiographic distribution of serotonin transporters and receptor subtypes in human brain. Hum Brain Mapp 22:246-260.
Woods RP, Cherry SR, Mazziotta JC (1992) Rapid automated algorithm for aligning and reslicing PET images. J Comput Assist Tomogr 16:620-633.
Contacts
For questions related to the NRU serotonin atlas, contact Vincent Beliveau or Melanie Ganz-Benjaminsen.
Copyright
Copyright © 2021, Neurobiology Research Unit, Rigshospitalet. All rights reserved according to CC 4.0 BY-NC-SA (https://creativecommons.org/licenses/by-nc-sa/4.0). Redistribution and use in source forms, with or without modification, are permitted provided that the following conditions are met: * You must give appropriate credit, provide a link to the license, and indicate if changes were made. You may do so in any reasonable manner, but not in any way that suggests the licensor endorses you or your use. * You may not use the material for commercial purposes. Neither the name of the Neurobiology Research Unit, Rigshospitalet nor the names of its contributors may be used to endorse or promote products derived from this image set without specific prior written permission. * If you remix, transform, or build upon the material, you must distribute your contributions under the same license as the original.
Files
The following files are available for download if you go one level up (https://nru.dk/FS5ht-atlas/):
| Size | File | Description |
| 4 kB | bmax_table.csv | Table with Bmax values for all regions, cf. Fig 3 |
| 16,4 MB | 5-HT_atlas.zip | Average density maps presented in Figs 1 and 2 and table for the data from Fig 3 |
| 825 kB | 5-HT_parcellation.zip | Clustering maps presented in Fig 4 |
Description
DATquan is a Matlab-based clinical software tool which can provide fast, accurate, and highly reproducible quantification of DAT binding in brain SPECT images.
The software is described in details in:
Jensen PS, Thomsen G, Ziebell M, deNijs, R, Svarer C, Malik U, Skouboe G, Knudsen GM. Validation of a Method for Accurate and Highly Reproducible Quantification of Brain Dopamine Transporter SPECT Studies. J. Nuclear Medicine and Technology2011;39(4):271-8
File
The following file is available for download below:
| Size | File | Description |
| 12,9 MB | DATquan.zip | Package with all needed files (Updated 28th Apr, 2020) |
Description
BBTools is a Matlab-toolbox for large-scale computations with a black-box operator. A black-box operator is an operator that can be accessed only through a pair of routines that compute matrix-vector multiplications.
At NRU we use this for research in new reconstruction algorithms for PET and SPECT. However, the toolbox should be useful for many other fields such as inverse problems in general, and dimensionality reduction of large data sets (e.g. latent semantic indexing).
Online documentation
You can see a web-version of the online documentation here. Please note that the documentation is designed to be used from Matlab. The online version is only an approximation to the experience.
Platforms
BBTools will now support the AMD64 architecture under Linux. Although untested, AMD-clones by Intel should also be supported. Thus the following platforms are supported out of the box:
- Windows/PC (64-bit is not yet native)
- Linux/PC (32- and 64-bit native)
- HP/HPUX
Funding
The development of the toolbox would not be possible without the financial support from the following organizations:
- EU 6th Framework Programme
Diagnostic Molecular Imaging
Contract LSHB-CT-2005-512146 (DiMI) - EU Fifth Framework Programme
Neuroreceptor Changes in Mild Cognitive Impairment
Contract QLK6-CT-2000-00502 (NCI-MCI) - M.L. Jørgensens og Gunnar Hansens Fond
J. nr. 2956 - Savværksejer Jeppe Juhl og hustru Ovita Juhls Mindelegat
Also, parts of the toolbox were developed at Tufts University, Massachussets, USA. This trip was made possible with funding from:
- Otto Mønsteds Fond
J. nr. 03-71-898
Bugs
A critical bug was fixed which made BBTools err when estimating the norm of a Matlab-matrix under some circumstances where some columns was zero. This, in turn, caused BBMATRIX to fail for the same class of matrices.
All users are encouraged to upgrade to at least version 0.2.4c, released 09-Nov-2005.
An issue has been found with the installer running under the Ubuntu variant of Linux. While BBTools will still work, the documentation is not properly integrated into the Matlab environment. As a temporary workaround, while this issue is being investigated, is to use the command BBDOC to read the online documentation.
Files
The following files are available for download below:
| Size | File | Description |
| 66,4 KB | bbinstall.m | Install, uninstall, upgrade, and download |
| 3,4 MB | bbtools.zip | The BBTools package |
File
The software can be downloaded from here. But first you must register at https://nru.dk/pveout in order to get download and installation instructions.
Description
Matlab-based software that has been developed in the EU sponsored PVEOut project. This software package includes everything that is needed to do a partial volume correction of low resolution medical images. An easy to use user interface is included. Further, multiple methods for labeling of voxels (defining regions), including the probability based method by Svarer et al (2005), Neuroimage 24, 969-79 (applyrois) is incorporated in the software.
Description
The dorsal raphe (DR) and median raphe (MR) masks were delineated in subject PET space as described in "Beliveau V, Svarer C, Frokjaer VG, Knudsen GM, Greve DN, Fisher PM. Functional connectivity of the dorsal and median raphe nuclei at rest. Neuroimage. 2015 Aug 1;116:187-95", then normalized to the MNI152 space (ICBM) to produce a probability map across subjects.
To find the transformation to normal space, PET and MR were coregistered using bbregister and then the MR was registered to MNI152 using flirt+fnirt in FSL. The two transforms were then concatenated and the masks were transferred to MNI152 space using nearest neighbor interpolation; these masks are the ICBM152.fnirt. However, fnirt is sometimes imprecise to align brainstem correctly and an alternative procedure was proposed by "Napadow V, Dhond R, Kennedy D, Hui K, Makris N. Automated brainstem co-registration (ABC) for MRI. Neuroimage. 2006; 32:1113-1119".
Essentially, it is a first alignment with flirt followed by a second flirt alignment focusing on brainstem by using a weighing limited to that region. We have applied that method and got slightly different masks, although nothing drastic. In our opinion, this second (ICBM152.brainstem.flirt) method yields better results, but feel free to use whichever you like.
These masks have also been included in the latest version of the AAL atlas: 10.1016/j.neuroimage.2019.116189.
Contacts
For questions related to the DR/MR masks, contact Vincent Beliveau.
Copyright
Copyright © 2021, Neurobiology Research Unit, Rigshospitalet.
Files
The following files are available for download if you go one level up:
| Size | File | Description |
| 4 kB | raphe_masks.zip | Zipped file containing the DR/MR masks |
Overview
Gamma-aminobutyric acid (GABA) is the main inhibitory neurotransmitter in the human brain and plays a key role in several brain functions and neuropsychiatric disorders such as anxiety, epilepsy, and depression. The binding of benzodiazepines to the benzodiazepine receptor sites (BZR) located on GABAA receptors (GABAARs) potentiates the inhibitory effect of GABA leading to the anxiolytic, anticonvulsant and sedative effects used for treatment of those disorders.
However, the function of GABAARs and the expression of BZR protein is largely determined by each GABAAR subunit stoichiometry (19 genes coding for individual subunits), and it is not well understood how these mechanisms may vary between individuals and across different brain regions.
Here, we present a quantitative high-resolution in vivo atlas of the human brain BZRs, generated on the basis of [11C]flumazenil Positron Emission Tomography (PET). Next, based on autoradiography data, we transform the PET-generated atlas from binding values into BZR protein density. Finally, we examine the brain regional association with mRNA expression for the 19 subunits in the GABAAR, including an estimation of the minimally required expression of mRNA levels for each subunit to translate into BZR protein.
This represents the first publicly available quantitative high-resolution in vivo atlas of the spatial distribution of BZR densities in the healthy human brain. The atlas provides a unique neuroscientific tool as well as novel insights into the association between mRNA expression for individual subunits in the GABAAR and the BZR density at each location in the brain.
Data
All participants included in this study were healthy controls from the Cimbi database (Knudsen et al., 2015); the data analysis was restricted to include individuals aged between 18 and 45 years. Participants were recruited by advertisement for different research protocols approved by the Ethics Committee of Copenhagen and Frederiksberg, Denmark. A total of 26 PET scans and corresponding structural MRI were acquired for 16 individual participants; 7 subjects had only 1 scan, 8 subjects had 2 scans, and a single had 3 scans.
Methods
PET data was acquired in list-mode on a Siemens HRRT scanner operating in 3D-acquisition mode, with an approximate in-plane resolution of 2 mm (1.4 mm in the center of the field of view and 2.4 mm in cortex) (Olesen et al., 2009). The radioligand was given either as a bolus or as a bolus-infusion protocol. PET frames were reconstructed using a 3D-OSEM-PSF algorithm (Comtat et al., 2008; Sureau et al., 2008). Scan time was 90 minutes divided into 35 frames (6x5, 10x15, 4x30, 5x120, 5x300, 5x600 seconds). Dynamic PET frames were realigned using AIR 5.2.5 (Woods et al., 1992). The PET data was quantified to estimate total distribution volumes (VT) for each brain region using steady-state analysis for the bolus-infusion protocol and Logan analysis for the bolus protocol, corrected for radio metabolites (Feng et al. 2016).
T1-weighted structural MRI were acquired on two different 3T Siemens scanners with standard parameters. For further details on structural MRI acquisition parameters, see Knudsen et al. (2015).
Please see Figure 1 for a full overview of the methodology.
Figure 1: Flowchart of the processing of the MRI and PET data using the radioligand [11C]Flumazenil, ranging from motion correction, matching of structural MRI using FreeSurfer (v. 6.0), kinetic modeling with arterial sampling, and finally establishing the association between postmortem human brain autoradiography from Braestrup et al. 1977 and the regional total distribution volumes (VT).
Atlas maps
The BZR density (pmol per gram protein) was obtained by normalizing VT with the corresponding postmortem human brain [3H]diazepam autoradiography data from Braestrup et al. 1977. The non-displaceable distribution volume (VND) was estimated as the intercept (Figure 2B). The atlas was transformed to represent protein densities in pmol/ml (Figure 2A). Finally, the association between protein density and mRNA expression for the 19 GABAAR subunits was assessed using the Allen Human Brain atlas (Hawrylycz et al. 2012). The association between the BZR density and the three commonly expressed subunits (a1, b2, and g2) can be found in Figure 3.

Figure 2: (A) High resolution atlas of GABAAR density (pmol/ml) in MNI152 space (left) and in fsaverage space (right) (B) Average regional distribution volumes (VT) and benzodiazepine receptor density for the GABAAR. The regional VT's determined by PET were matched to the corresponding regions from the [3H]diazepam autoradiography data. The regression is shown as the black line, and the intercept is the non-displaceable distribution volume (VND). The shaded area is the 95% confidence interval.

Figure 3: (A-C) Association between mRNA expression (log2 intensity) and BZR density for the subunits of the GABAAR most commonly represented in the BZR, a1 in (A), b2 in (B), and g2 in (C). The points are regional estimates for subcortex (squares) and cortex (round dots), and are color coded according to the density, green (<800 pmol/ml), blue (800-900 pmol/ml), red (900-1.000 pmol/ml), and black (>1.000 pmol/ml). (D) Biplot of the (scaled) first two principal components (% variance explained) of a PCA of the 19 subunits and BZR. (E-F) The spatial distribution of BZR density according to the specified color coding shown on the lateral and medial surface of the brain.
Publications
The following publication should be referenced when using this atlas:
Nørgaard M, Beliveau V, Ganz M, Svarer C, Pinborg LH, Keller SH, Jensen PS, Greve DN, Knudsen GM. A high-resolution in vivo atlas of the human brain's benzodiazepine binding site of GABAA receptors. Neuroimage. 2021 May 15;232:117878. doi: 10.1016/j.neuroimage.2021.117878.
Downloads
All the atlas data, and the supplementary data can be found here (9.3 MB).
Bibliography
Braestrup, C., Albrechtsen, R., & Squires, R. (1977). High densities of benzodiazepine receptors in human cortical areas. Nature, 269(October), 702-704.
Comtat C, Sureau FC, Sibomana M, Hong IK, Sjoholm N, Trebossen R (2008) Image based resolution modeling for the HRRT OSEM reconstructions software. In: 2008 IEEE Nuclear Science Symposium Conference Record, pp 4120-4123. IEEE.
Feng, L., Svarer, C., Madsen, K., Ziebell, M., Dyssegaard, A., Ettrup, A., Pinborg, L. H. (2016). Design of Infusion Schemes for Neuroreceptor Imaging: Application to [11C]Flumazenil-PET Steady-State Study. BioMed Research International, 2016.
Hawrylycz, M. J., Lein, E. S., Guillozet-Bongaarts, A. L., Shen, E. H., Ng, L., Miller, J. A., Jones, A. R. (2012). An anatomically comprehensive atlas of the adult human brain transcriptome. Nature, 489(7416), 391-399.
Knudsen GM et al. (2015) The Center for Integrated Molecular Brain Imaging (Cimbi) Database. Neuroimage:1-7.
Olesen OV, Sibomana M, Keller SH, Andersen F, Jensen J, Holm S, Svarer C, Hojgaard L (2009) Spatial resolution of the HRRT PET scanner using 3D-OSEM PSF reconstruction. IEEE Nucl Sci Symp Conf Rec:3789-3790.
Sureau FC, Reader AJ, Comtat C, Leroy C, Ribeiro M-J, Buvat I, Trebossen R (2008) Impact of image-space resolution modeling for studies with the high-resolution research tomograph. J Nucl Med 49:1000-1008.
Woods RP, Cherry SR, Mazziotta JC (1992) Rapid automated algorithm for aligning and reslicing PET images. J Comput Assist Tomogr 16:620-633.
Contacts
For questions related to the NRU BZR atlas, contact Martin Norgaard or Gitte M. Knudsen.
Copyright
Copyright (c) 2016, Neurobiology Research Unit, Rigshospital. All rights reserved according to CC 4.0 BY-NC-SA (https://creativecommons.org/licenses/by-nc-sa/4.0).

















